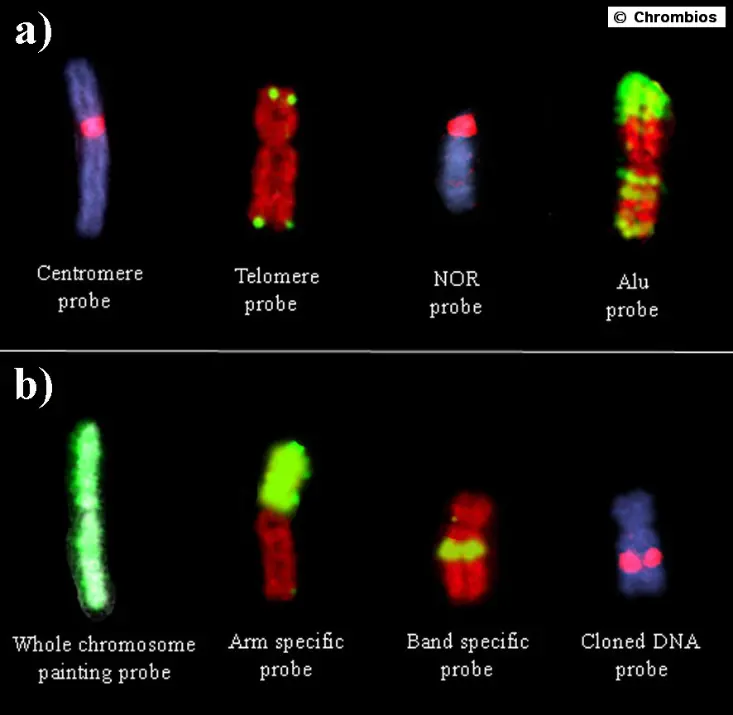
Repetitive probes
Centromere repeats
Telomere repeats
Ribosomal DNA (rDNA)
Interspersed repetitive elements (SINEs, LINEs)
The first in situ hybridization experiments to chromosomes were performed with highly repetitive DNA sequences from mouse centromeres. Centromeres and telomeres of mammalian chromosomes usually contain large arrays of simple DNA motives that produce strong signals with FISH (Figure 4 a, b). Although telomere sequences are highly conserved during evolution, the centromeric repeats may evolve rather fast. Closely each centromere of the human karyotype can be differentiated by FISH with specific probes (satellite and alphoid DNAs). Even within the centromere distinct blocks of different repeats can be observed in various chromosomes. Probes derived from these repeats provide a valuable tool for the identification of chromosomes and for interphase FISH.
Another type of clustered repeats are the ribosomal DNA sequences (rDNA) that are found close to the centromeres of human chromosome 13-15, 21 and 22 and form the "nucleolar organizing regions" (NOR) (Figure 4 c).
Another group of repeated sequences are dispersed all over the genome such as "short interspersed elements" (SINEs) and "long interspersed elements" (LINEs). These sequences are not evenly distributed over the chromosomes but seem to be more concentrated in certain chromosome bands, as is found for the Alu-elements that are more concentrated in the R-bands (Figure 4 d). Thus, FISH with Alu sequences will result in an R-band like pattern and can be used for chromosome identification.