Chromosome bar codes
Cross-species color segmenting or RxFISH
Bar codes from fragmented hybrids
Multi color banding
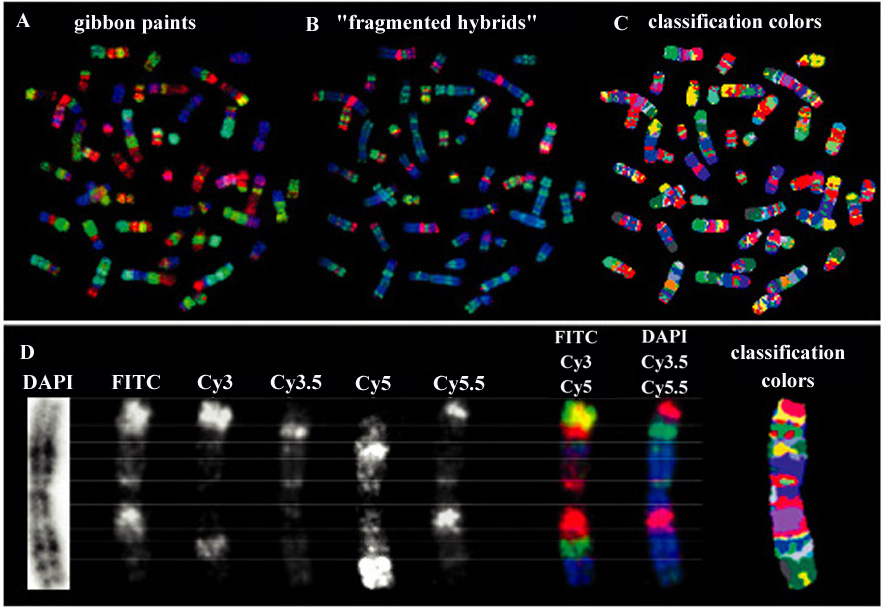
Chromosome painting allows the identification of whole chromosomes only and therefore is limited to the analysis of chromosome translocations. The delineation of intra-chromosomal rearrangements (inversions, duplications, transpositions, etc.) requires sub-regional definition of the chromosomes preferentially by molecular probes. FISH has been used to produce a highly defined colored banding pattern of chromosomes in a multi-color format which was named "chromosome bar coding" (Figure 18).
Two alternative strategies for a "bar code" based differentiation of the entire human karyotype with molecular probes have been proposed. Both approaches make use of multi color FISH with probes derived from fragmented chromosomes that would paint sub-regions or bands in the respective colors.
Cross-species color segmenting or RxFISH
Since gibbon (hominoid primates) karyotypes have been highly fragmented by translocations during evolution, painting probes derived from these species can serve to delineate up to 100 different chromosome segments in the human karyotype (cross-species color segmenting or RxFISH, (Figure 18a).
Bar codes from fragmented hybrids
The other bar code approach makes use of fragmented chromosomes in human/rodent somatic cell hybrids (Figure 18b). Human DNA can be seperated from rodent DNA by PCR using primers specific for Alu repeats which are confined to primates. Using this approach human chromosome segment specific painting probes from these cell lines were generated. A large number of rearranged hybrid lines containing more than 300 human chromosome fragments have been characterized by in situ hybridization to normal human metaphase chromosomes. Various of these "fragmented hybrids" were used to produce a chromosome bar code with up to 110 molecular landmarks on human chromosomes.
Both "bar codes" can also be co-hybridized to human chromosomes in a single FISH experiment employing six different fluorochromes (Figure 18d). Together, both bar codes yield approximately 160 molecular landmarks in multiple colors, that would correspond to a resolution of about 400 bands with classical banding techniques.
Multi color banding (MCB)
Another approach to introduce colored patterns on chromosomes by FISH is using multi color FISH with band specific probes derived from micro-dissection of chromosome bands.This approach was used to colorize individual chromosomes but not entire karyotypes.
Chromosome bar coding has been used to study various solid tumors and haematological malignancies.The genome-wide screening technique was used as a complement to G-banding in cases chromosomal rearrangements involved segments too small, too similarly banded, or too complex to be described adequately or even to be detected by G-banding.